
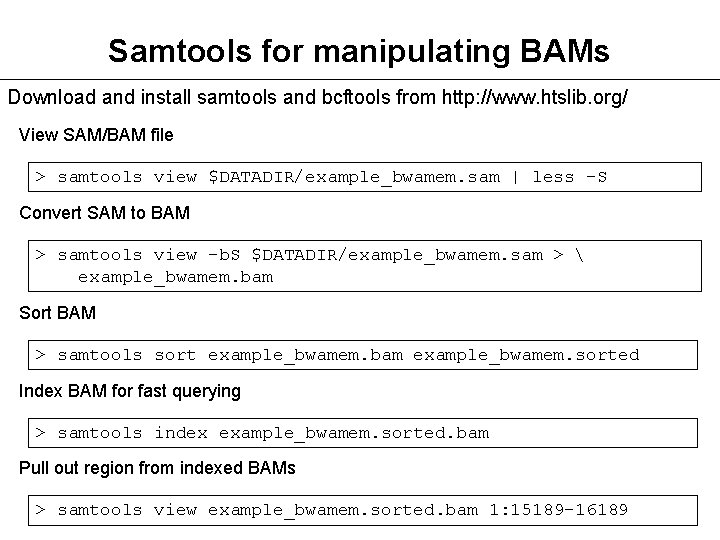
A useful command is view which converts a BAM file to SAM. configure -prefix/where/to/install make make install See INSTALL in each of the source directories for further details.

Here are three of my favorite Python Bioinformatics Books in case you want to learn more about it. All you need to do is to use the command lines above, but replace the sub command fasta for fastq. samtools: packagesamtools0119 packagesamtools11 packagesamtools12 I used toolshed.g2.bx.psu.edu suitsamtools12 and dont know why all the versions of samtools are installed. samtools also has a mode to convert from BAM to FASTQ. samtools command -paramter -parameter bam/sam-file. Building and installing Building each desired package from source is very simple: cd samtools-1.x and similarly for bcftools and htslib. Using Samtools to Convert a BAM into FASTQ. The main samtools source code repository moved to GitHub in March 2012. SAMtools provide efficient utilities on manipulating alignments in the SAM format. The basic pattern of usage for samtools is. SAM (Sequence Alignment/Map) is a flexible generic format for storing nucleotide sequence alignment. The command man samtools shows you a longer documentation. We recommend using anaconda and the conda command to install dependenciesĪccept all defaults for “Do you wish the installer to initialize Anaconda3 by running conda init?” say Yes. You can run samtools without any parameters to get an overview of parameters and options. For more information on how to download earlier versions of samtools git repositories or stm, bcftools, or htslib releases) or from the samtools Sourceforge project. To run VeloCyto, you will need Python >=3.6.0 (we have no plans to support python<=3.5). Is Htslib Installed also available as a separate package that is suitable for developing tools against it by writing your own programs.


 0 kommentar(er)
0 kommentar(er)
